| Cardiology Research, ISSN 1923-2829 print, 1923-2837 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Cardiol Res and Elmer Press Inc |
| Journal website https://www.cardiologyres.org |
Original Article
Volume 15, Number 4, August 2024, pages 262-274
FASLG as a Key Member of Necroptosis Participats in Acute Myocardial Infarction by Regulating Immune Infiltration
Hui Min Jiaa, b, Fu Xiang Ana, b, Yu Zhanga, Mei Zhu Yana, Yi Zhoua, Hong Jun Biana, c
aDepartment of Emergency Medicine, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong 250021, China
bThese authors contributed equally to this work.
cCorresponding Author: Hong Jun Bian, Department of Emergency Medicine, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong 250021, China
Manuscript submitted April 26, 2024, accepted July 17, 2024, published online July 30, 2024
Short title: FASLG is a Regulator in AMI
doi: https://doi.org/10.14740/cr1652
| Abstract | ▴Top |
Background: Acute myocardial infarction (AMI) is a major cause of human health risk. Necroptosis is a newly and recently reported mode of cell death, whose role in AMI has not been fully elucidated. This study aimed to search for necroptosis biomarkers associated with the occurrence of AMI and to explore their possible molecular mechanisms through bioinformatics analysis.
Methods: The dataset GSE48060 was used to perform weighted gene co-expression network analysis (WGCNA) and differential analysis. Key modules, differential genes, and necroptosis-related genes (NRGs) were intersected to obtain candidate biomarkers. Groups were classified and differentially analyzed according to the expression of the key biomarker. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, gene set enrichment analysis (GSEA), and construction of protein-protein interaction (PPI) networks are performed on differentially expressed genes (DEGs). Finally, CIBERSORT was used to assess immune cell infiltration in AMI and the correlation of key biomarkers with immune cells. Immune cell infiltration analysis revealed the correlation between FASLG and multiple screened immune cells.
Results: WGCNA determined that the MEsaddlebrown module was the most significantly associated with AMI. Intersecting it with DEGs as well as NRGs, we obtained two key genes, FASLG and IFNG. But only FASLG showed statistically significant differences between the AMI group and the normal control group. Further analysis suggested that the down-regulation of FASLG may exert its function through the regulation of the central genes CD247 and YES1. Furthermore, FASLG was positively correlated with T-cell CD4 memory activation and T-cell gamma delta, and negatively correlated with macrophage M0.
Conclusion: In conclusion, FASLG and its regulatory genes CD247 and YES1 might be involved in the development of AMI by regulating immune cell infiltration. FASLG might be a potential biomarker for AMI and provides a new direction for the diagnosis of AMI.
Keywords: Acute myocardial infarction; Necroptosis; FASLG; Immune infiltration; Bioinformatics
| Introduction | ▴Top |
The latest data from the American Heart Association revealed that the crude incidence of cardiovascular disease (CVD) increased by 29.01% and the mortality rate increased by 18.71% in 2020 compared to 2010. CVD continued to be the leading cause of death worldwide. Acute myocardial infarction (AMI), characterized by interruption of blood flow to the heart, is a common CVD [1]. The age of onset of AMI is becoming younger and younger in recent years, making it a major risk factor for human health in all age groups [2]. The diagnosis of AMI depends mainly on the electrocardiogram and changes in myocardial injury markers (e.g., troponin T, creatine kinase isoenzymes, myoglobin, etc.) [3]. However, the sensitivity and specificity of currently used markers of myocardial injury are still unsatisfactory [4, 5]. Therefore, exploring new blood biomarkers is particularly important for the diagnosis of AMI.
With the development of high-throughput sequencing technology and bioinformatics in recent years, new diagnostic and prognostic biomarkers are increasingly being discovered. It is generally known that primary lesion tissue for CVD is difficult to be obtained. It has been observed that the peripheral blood transcriptome has an 80% overlap with genes expressed in human tissues such as the heart [6]. The peripheral blood gene expression profile has been validated in a variety of diseases involving inflammation, and the role of inflammation in CVDs such as AMI has also been demonstrated. This relationship makes peripheral blood an easily available sample to explore the disease process of AMI and seems to be a viable alternative [7]. Applying peripheral blood bioinformatics analysis, it has been identified that leukocyte immunoglobulin-like receptor B2 (LILRB2), neutrophil cytosolic factor 2 (NCF2), Toll-like receptor 2 (TLR2), Fc fragment of IgE receptor Ig (FCER1G), formyl peptide receptor 1 (FPR1), etc. may be potential biomarkers for the diagnosis and treatment of AMI [8-10]. Despite these advances, the critical biomarkers that serve an important role in AMI have remained incompletely explored.
Necroptosis is a newly discovered mode of cell death that is characterized by enhanced plasma membrane permeability and mitochondrial swelling [11]. In necroptotic apoptosis, receptor-interacting protein kinase-3 (RIPK3) is recruited to necrotic vesicles phosphorylated by receptor-interacting protein 1 (RIP1). Activated RIPK3 then phosphorylates the mixed lineage kinase domain-like protein (MLKL) and generates a pore complex at the plasma membrane. This pore complex disrupts the integrity of the plasma membrane and increases its permeability, causing an inward flow of Ca2+ and Na+, releasing cellular contents such as mitochondrial DNA, and ultimately leading to cell swelling and membrane rupture [12]. Necroptosis has been identified to function in a variety of diseases, such as atherosclerosis, myocardial infarction, ischemia/reperfusion (I/R) injury, chronic intestinal inflammation, and tumors [13-17]. In recent years, several studies have revealed the relationship between necroptosis-related genes (NRGs) and AMI, such as pigment epithelium-derived factor (PEDF) and its functional peptides protect cardiomyocytes from hypoxia-induced necroptosis in AMI [18]; TNFR1 and DR6 serve as direct targets of miR-223-5p and are engaged in myocardial I/R-induced inflammation and necroptosis [19]. However, the roles that NRGs play in AMI have not been fully elucidated.
In this study, we performed weighted gene co-expression network analysis (WGCNA) on the dataset GSE48060 to identify key gene co-expression modules and analyzed them bioinformatically. We found that the NRG FASLG has reduced expression after AMI and may be a potential biomarker of myocardial infarction. Furthermore, we grouped FASLG with differential expression and explored the possible mechanisms of FASLG involvement in myocardial infarction.
| Materials and Methods | ▴Top |
Data download and preprocessing
Genome-wide microarray datasets GSE48060 and GSE60993 were obtained from the Gene Expression Omnibus (GEO) [20]. GSE48060 was based on the GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array platform. This dataset contained 52 human peripheral blood samples, including 31 blood samples from patients with their first AMI within 48 h of onset and 21 blood samples from normal heart function controls. GSE60993 was based on the GPL6884 Illumina HumanWG-6 v3.0 expression beadchip platform and included 17 patients with AMI, nine patients with unstable angina pectoris, and seven healthy controls. The study sample consisted of whole blood collected from patients with a first AMI within 48 h of MI and controls with normal echocardiograms. Patients with prior participation in a cardiac rehabilitation programme, a history of CVD, or clinical or biochemical evidence of other comorbidities such as cancer, rheumatoid arthritis, liver disease, or myeloproliferative disorders were excluded. For samples in the GSE48060 and GSE60993 datasets, written informed consent was obtained from all subjects and approved by the Ethics Committee [21, 22]. GSE48060 was adopted as the experimental set and GSE60993 as the validation set. Probes were converted to gene symbols using the R package. If a plurality of probes matched a gene symbol, the first occurring expression value was selected as the expression value for that gene. Meanwhile, we collected 159 human NRGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) [23].
WGCNA
WGCNA was designed to search for co-expressed gene modules and to explore the association between gene networks and phenotypes of interest, as well as the core genes in the network. We executed WGCNA on all samples in the GSE48060 dataset using the WGCNA package in R software to identify key modules. The “pickSoftThreshold” function calculated a weighted network by the expression similarity of all genes. It analyzed information such as the scale-free topological fit index and connectivity of the network at the specified soft thresholding power, to assist in the selection of a suitable soft thresholding power for network construction. Subsequently, a scale-free network was constructed using selected soft thresholding power. The similarity of expression between genes was calculated based on the gene expression value matrix to obtain the neighborhood matrix and then computed the topological overlap matrix. Co-expression modules were further identified by drawing hierarchical clustering trees based on the topological overlap matrix. The minimum number of genes per gene module was set at 30 and similar modules merged using 0.25 as the cut height. Correlation coefficients between co-expression modules and diseases were calculated and the modules with the highest correlation coefficients and the genes they contained were selected for subsequent analysis.
Identification of key genes
The R package “FactoMineR” was used to perform principal component analysis (PCA) on the AMI and healthy control samples, and the R package “factoextra” was used for visualization. Differentially expressed genes (DEGs) were selected using the R package “limma” and the selection criterion was set to be |logFC| > 0.263, P value < 0.05. The DEGs, the genes in the key modules of the WGCNA, and the NRGs were then intersected to obtain possible biomarkers, using the R package “ggvenn” to validate them as Venn diagrams. To further identify key genes, a Wilcox rank-sum test was used to verify whether the expression of the intersecting genes differed between the AMI and healthy controls. Construct a receiver operating characteristic (ROC) curve to determine the diagnostic value and validate it in GSE60993. These analyses were performed using the “wilcox. test” function and the R package “ROCR”. FASLG was chosen as the key gene for subsequent analysis based on the foregoing analysis.
Analysis of DEG identification and functional enrichment
The AMI samples in the GSE48060 dataset were divided into a high FASLG expression group and a low FASLG expression group according to their FASLG expression. PCA was performed for both groups using the R package “FactoMineR”, and the “limma” package was used to analyze the differences between the two groups, with the difference criterion set at |logFC| > 0.263, P value < 0.05.
Gene Ontology (GO) analysis annotates and classifies genes by biological pathway (BP), molecular function (MF), and cellular components (CC), and locates the most likely associated GO term for the differential gene by performing GO terms enrichment statistics on the differential gene. The KEGG database is a comprehensive database that integrates gene function, linked genomic information, and functional information, with a pathway database for storing information on gene pathways in different species. The R package “clusterProfiler” was used to implement GO and KEGG enrichment analysis and to visualize it using the “GOplot” package.
Gene set enrichment analysis (GSEA)
GSEA was used to estimate whether a predefined set of genes shows significant and consistent differences between two biological states. We used GSEA to analyze the enriched functions and pathways in the high and low FASLG expression groups, avoiding the omission of genes that are not significantly differentially expressed but are biologically important.
Construction of protein-protein interaction (PPI) network
The PPI network was constructed through the online analysis site STRING [24] database. The obtained protein interaction relationship data were imported into Cytoscape for visualization, and the top 10 core genes were filtered using the MCC method in the plug-in CytoHubba.
Next, we used the function “wilcox. test” to verify whether the expression of the core genes differed between the AMI group and the normal control group. Then we used the function “cor” to explore the correlation between the differentially expressed core genes and FASLG.
Analysis of immune cell infiltration assessment
The CIBERSORT algorithm evaluates the abundance of individual immune cell subpopulations using deconvolution. The R package “CIBERSORT” was used to calculate the composition ratio of 22 immune cell types. Two types of non-expressing immune cells were removed from the samples. Box plots were drawn using the “ggplot2” package to compare immune cell expression differences in the FASLG high expression group, FASLG low expression group, and normal control group. Correlation analysis was performed to explore the correlation between key genes and immune cells, and visualized by the R package “ggplot2”.
| Results | ▴Top |
One hundred nine genes in Saddlebrown Module were identified to be the most closely associated with the onset of AMI by WGCNA
All genes contained in the 52 samples in GSE48060 were analyzed by WGCNA to identify key modules and hub genes associated with AMI. Hierarchical clustering was conducted on all samples and no outliers were found (Fig. 1a). We selected β = 8 as the optimal soft threshold power, in which case the constructed network is more consistent with the scale-free network characteristics (Fig. 1b). A total of 14 modules were constructed after setting 0.25 as the merging threshold to merge similar modules (Fig. 1c), with each represented by one color (Fig. 1d). Genes that belonged to none of the other modules were included in the gray modules. Further analysis of the correlation between gene co-expression modules and clinical features revealed that the saddlebrown module was the most correlated module with AMI (P = 1 × 10-4) (Fig. 1e), and a total of 109 genes are contained in this module.
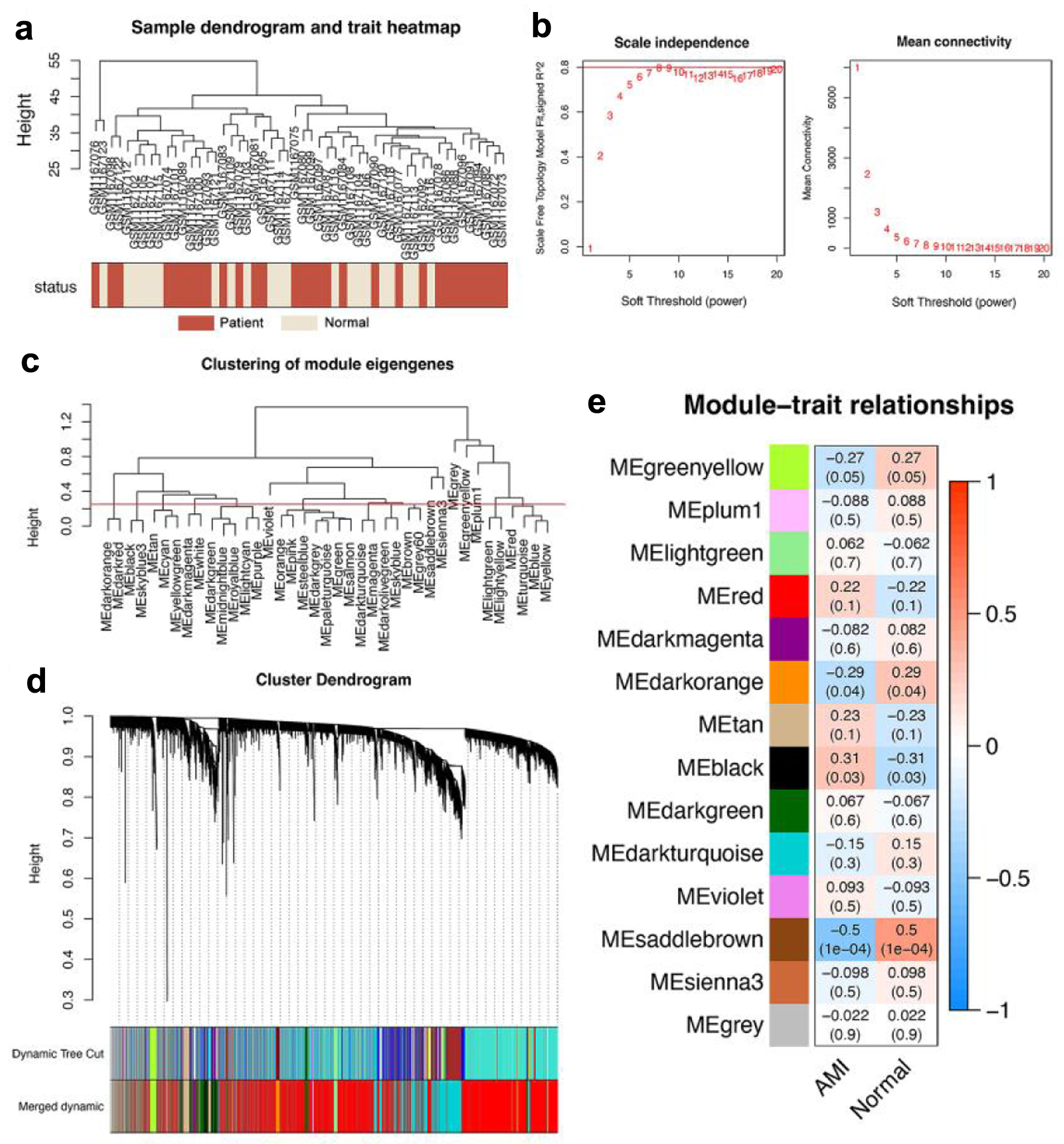 Click for large image | Figure 1. WGCNA revealed that genes contained in the saddlebrown module were maximally associated with AMI. (a) Cluster dendrogram of 52 samples, including 31 AMI samples (purplishred) and 21 normal control samples (lightyellow). (b) Analysis of network topology for various soft-thresholding powers, and 8 was the fittest power value. (c) Clustering diagram of module feature genes. (d) Clustering dendrogram of genes, with dissimilarity based on the topological overlap. A total of 14 modules were acquired after consolidation, and the saddlebrown module was considered to be the most relevant to AMI. (e) Heatmap showing the correlation between different color modules and clinical traits. AMI: acute myocardial infarction; WGCNA: weighted gene co-expression network analysis. |
FASLG was a key gene associated with necroptosis for AMI diagnosis
The PCA demonstrated differences in gene expression levels between the AMI and normal control groups in GSE48060 (Fig. 2a). We identified 598 DEGs by variance analysis, including 431 up-regulated genes and 167 down-regulated genes. DEGs were represented by volcano plots (Fig. 2b). After intersecting the DEGs with the genes in the brown module and the necrotizing apoptosis-related genes, we obtained two overlapping genes, FASLG and IFNG (Fig. 2c). We tentatively suggested that FASLG and IFNG are key necrotizing apoptotic genes associated with AMI. Both genes FASLG and IFNG were decreased in AMI samples compared with normal control samples; however, we found that down-regulation of FASLG was significant by the Wilcox rank-sum test (P = 1.89687 × 10-5), while down-regulation of IFNG was no significant difference by the same assay (P = 0.07021089 (Fig. 2d, e). Plotting the ROC curve evaluated the diagnostic ability of FASLG and IFNG for AMI. The area under the ROC curve (AUC) of FASLG reached 0.836, while the AUC of IFNG was only 0.650 (Fig. 2f, g). FASLG presented a significant difference in the test set GSE60993 and reached an AUC of 0.82, with good diagnostic performance (Fig. 2h, i). FASLG showed better diagnostic performance, therefore, FASLG was recommended as a hub gene to be investigated further.
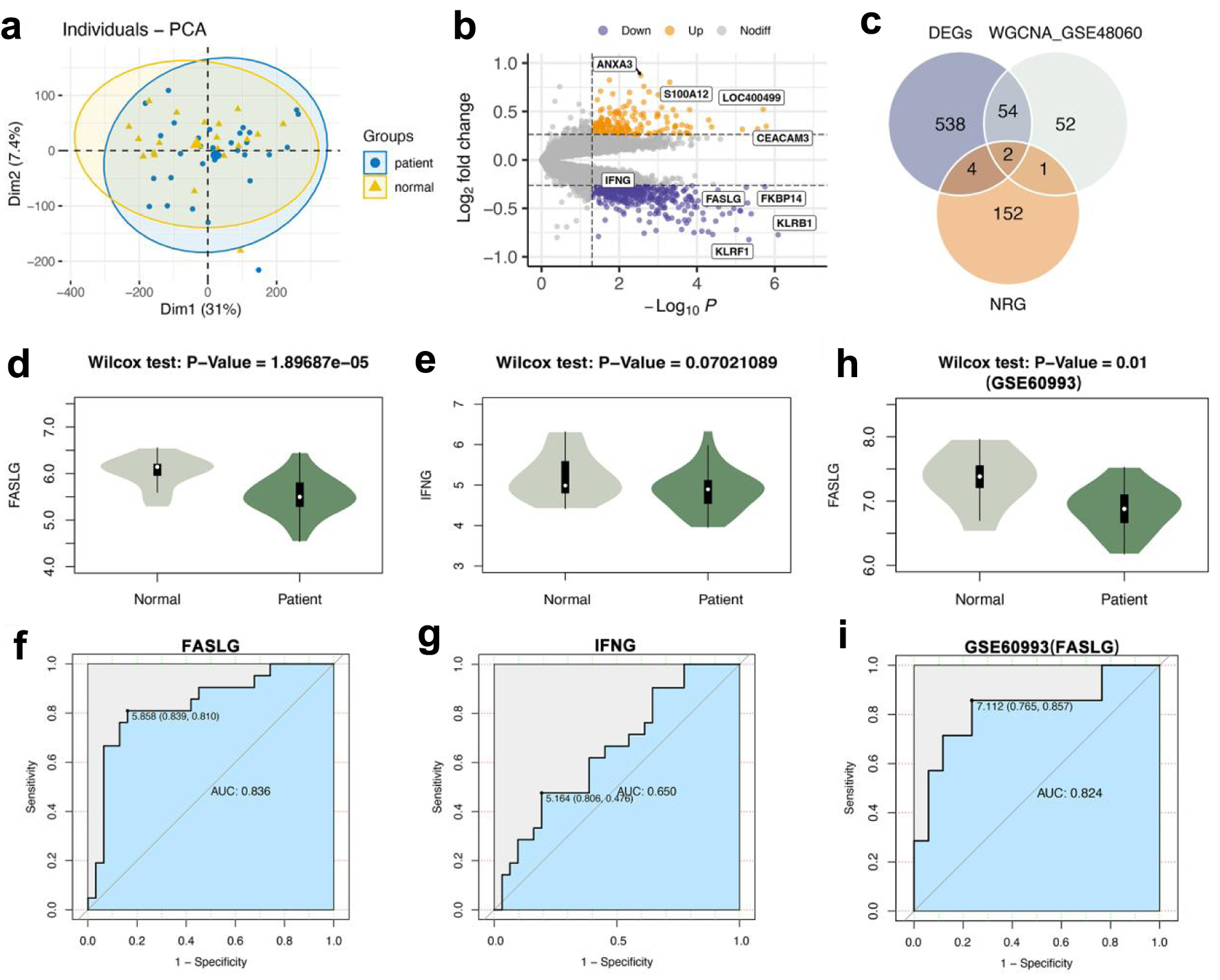 Click for large image | Figure 2. FASLG was a key gene associated with necroptosis for AMI diagnosis. (a) PCA of AMI group and normal control group. (b) Volcano plot of difference analysis between AMI group and normal group in GSE48060. (c) Venn diagram of intersecting genes in DEGs, WGCNA_GSE48060, NRG. (d) The Wilcox rank-sum test for the expression level of FASLG in the AMI and normal control groups (P = 1.89687 × 10-5). (e) The Wilcox rank-sum test for the expression level of IFNG in the AMI group and normal control group (P = 0.07021089). (f) The ROC curve of FASLG to differentiate the AMI group and normal control group with an AUC of 0.836. (g) The ROC curve of IFNG to differentiate the AMI group and normal control group with an AUC of 0.650. (h) The Wilcox rank-sum test for the expression level of FASLG in GSE60993 (P = 0.01). (i) The ROC curve of FASLG to differentiate the AMI group and normal control group in GSE60993 with an AUC of 0.824. AUC: area under the ROC curve; AMI: acute myocardial infarction; DEGs: differentially expressed genes; NRG: necroptosis-related gene; PCA: principal component analysis; ROC: receiver operating characteristic; WGCNA: weighted gene co-expression network analysis. |
Detection and enrichment analysis of differential genes in FASLG high and low expression groups of AMI patients
To explore the mechanism of FASLG involved in AMI regulation, the median FASLG expression was used as the boundary to divide the 31 AMI samples in GSE48060 into a high FASLG expression group and a low FASLG expression group. PCA indicated that the gene expression levels differed between the two groups (Fig. 3a). A total of 769 differential genes, including 506 down-regulated genes and 263 up-regulated genes, were obtained from differential analysis of the high and low expression groups of FASLG (Fig. 3b). These genes were considered as related genes of FASLG. A volcano plot was created and the top 100 genes with |logFC| values were selected for a heat map to visualize the results (Fig. 3c).
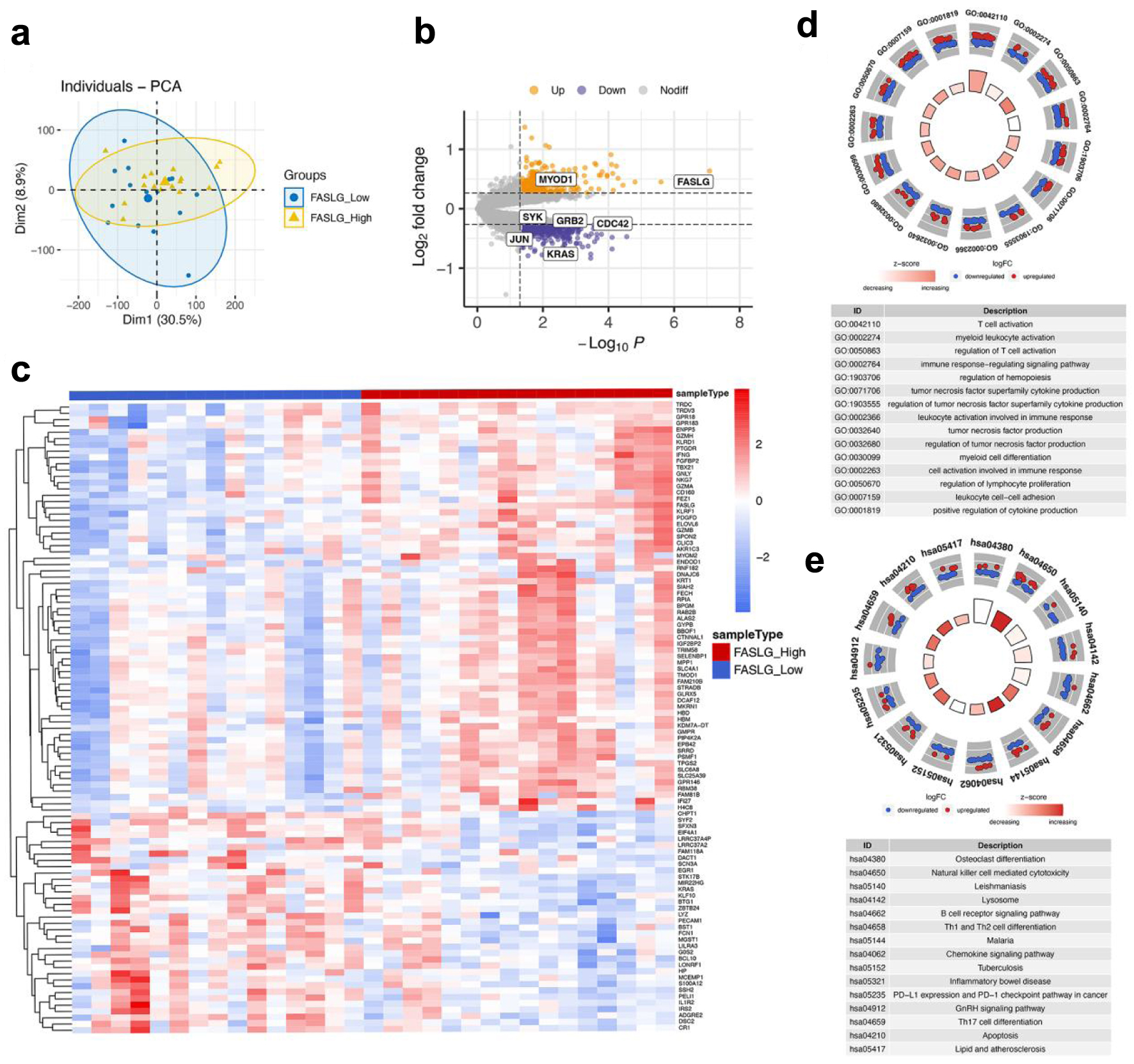 Click for large image | Figure 3. Differentially expressed gene identification and functional enrichment of FASLG high and low expression groups. (a) PCA was performed on the FASLG high expression and FAGLG low expression groups. (b) Volcano plot for differential analysis of FASLG high and low expression groups. Purple dots represent down-regulated genes and yellow dots represent up-regulated genes. (c) Heatmap showed the top 100 genes with |logFC| values among the differentially expressed genes in the FASLG high expression and FASLG low expression groups. (d) GO analysis of differentially expressed genes. (e) KEGG analysis of differentially expressed genes. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PCA: principal component analysis. |
In order to explore the function of FASLG, we further performed GO enrichment analysis and KEGG enrichment analysis on these differential genes. In the GO enrichment analysis, we concentrated on the BP part. The results show that these differential genes are mainly enriched in the GO terms of, for instance, regulation of T-cell activation, immune response-regulating signaling pathway, cell activation involved in immune response, and regulation of lymphocyte proliferation (Fig. 3d). Additionally, KEGG enrichment analysis demonstrated that differential expression of FASLG probably functioned through pathways such as natural killer cell-mediated cytotoxicity, B-cell receptor signaling pathways, Th1 and Th2 cell differentiation, Th17 cell differentiation, lipids and atherosclerosis, and apoptosis (Fig. 3e). The foregoing analysis indicated that FASLG might be involved in the progression of AMI by modulating immune cells and functions.
GSEA
We also performed a GSEA on the expression of all genes in the FASLG grouping to find consistently different enriched functions and pathways in the two groups to avoid the one-sidedness of the differential gene function enrichment analysis. Figure 4a, b illustrates the top 20 GO enrichment and KEGG enrichment entries, respectively. GO enrichment analysis showed that several biological processes were converged, for instance, the B-cell activation, the regulation of lymphocyte activation, the immune response-regulating signaling pathway, the positive regulation of cytokine production, and the Toll-like receptor signaling pathway (Fig. 4c). KEGG enrichment analysis revealed that low expression of FASLG may act through B-cell receptor signaling pathways, lipid and atherosclerosis, and oxidative phosphorylation (Fig. 4d). The conclusions drawn from the GSEA corroborated with the GO and KEGG enrichment analysis, further confirming the possible involvement of FASLG in the AMI process through the regulation of immune cells and functions.
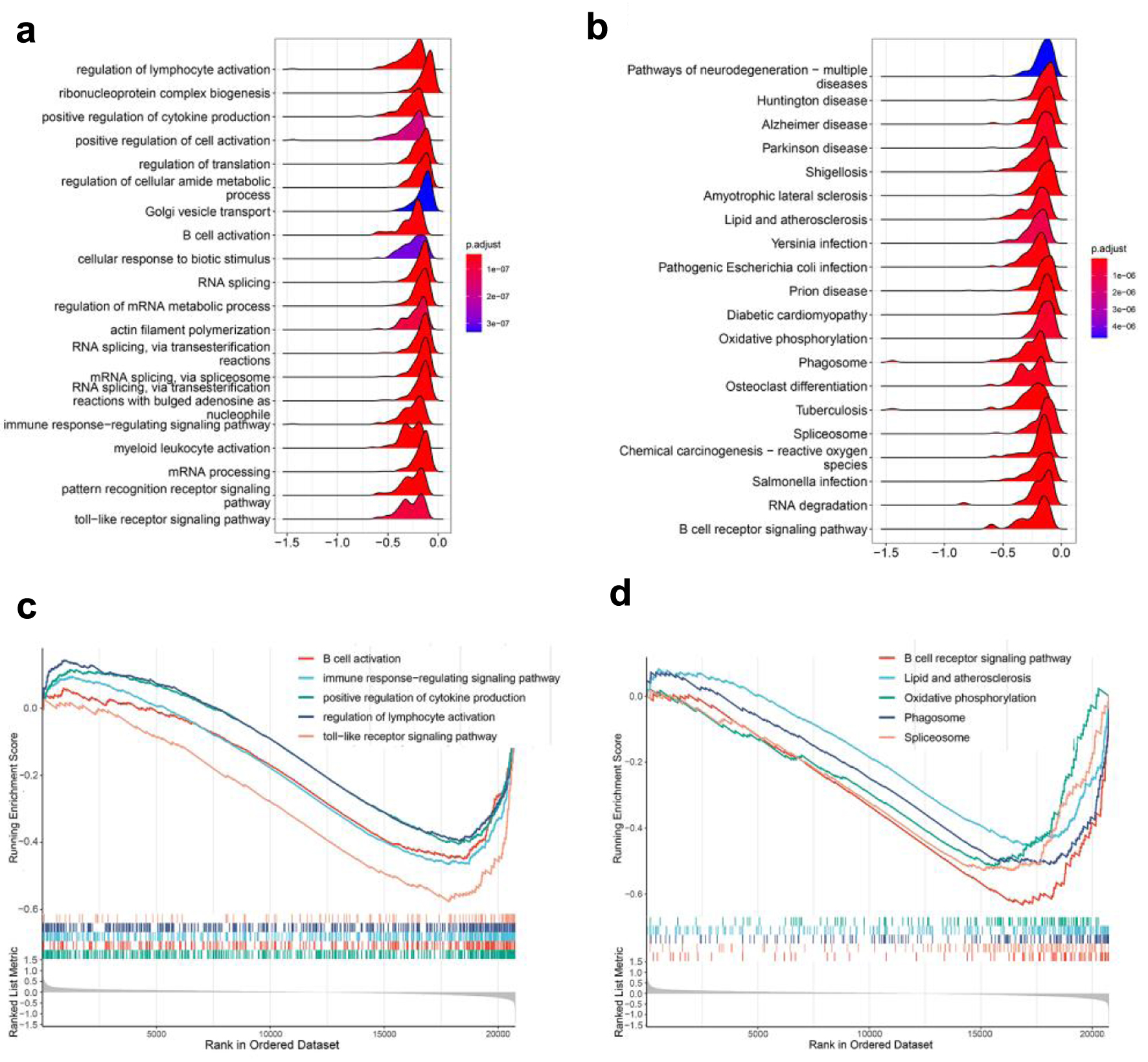 Click for large image | Figure 4. GSEA analysis. (a) GO enrichment analysis of GSEA enrichment analysis. (b) KEGG enrichment analysis of GSEA enrichment analysis. (c) A portion of the enriched GO terms. (d) A portion of the enriched KEGG pathways. GSEA: gene set enrichment analysis; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes. |
Construction of PPI networks and identification of hub genes based on FASLG
To further explore the potential mechanisms underlying the role of FASLG in AMI, we constructed a PPI network of FASLG-associated genes using the STRING database, which included 212 nodes and 405 edges (Fig. 5a). Ten central genes (LYN, YES1, SYK, JAK2, GRB2, HCK, FGR, CD247, BLK, and PRKCD) were identified by calculating MCC values through the CytoHubba plugin in Cytoscape (Fig. 5b). These 10 genes are those with a strong association with FASLG. Subsequently, our analysis revealed that the expression levels of YES1, HCK, CD247, and PRKCD in the AMI group and the healthy control group presented certain differences, in which the HCK and PRKCD expression was higher in the AMI group than in the control group, while YES1 and CD247 expression was lower in the AMI group than in the control group (Fig. 5c), and they may be equally involved in the development of AMI. The Pearson correlation analysis indicated that CD247 and YES1 presented a more significant positive correlation with FASLG expression. In contrast, the negative correlation between HCK, PRKCD, and FASLG expression was not significant (Fig. 5d). Therefore, CD247 and YES1 might be the underlying regulators of FASLG and are involved in the AMI process.
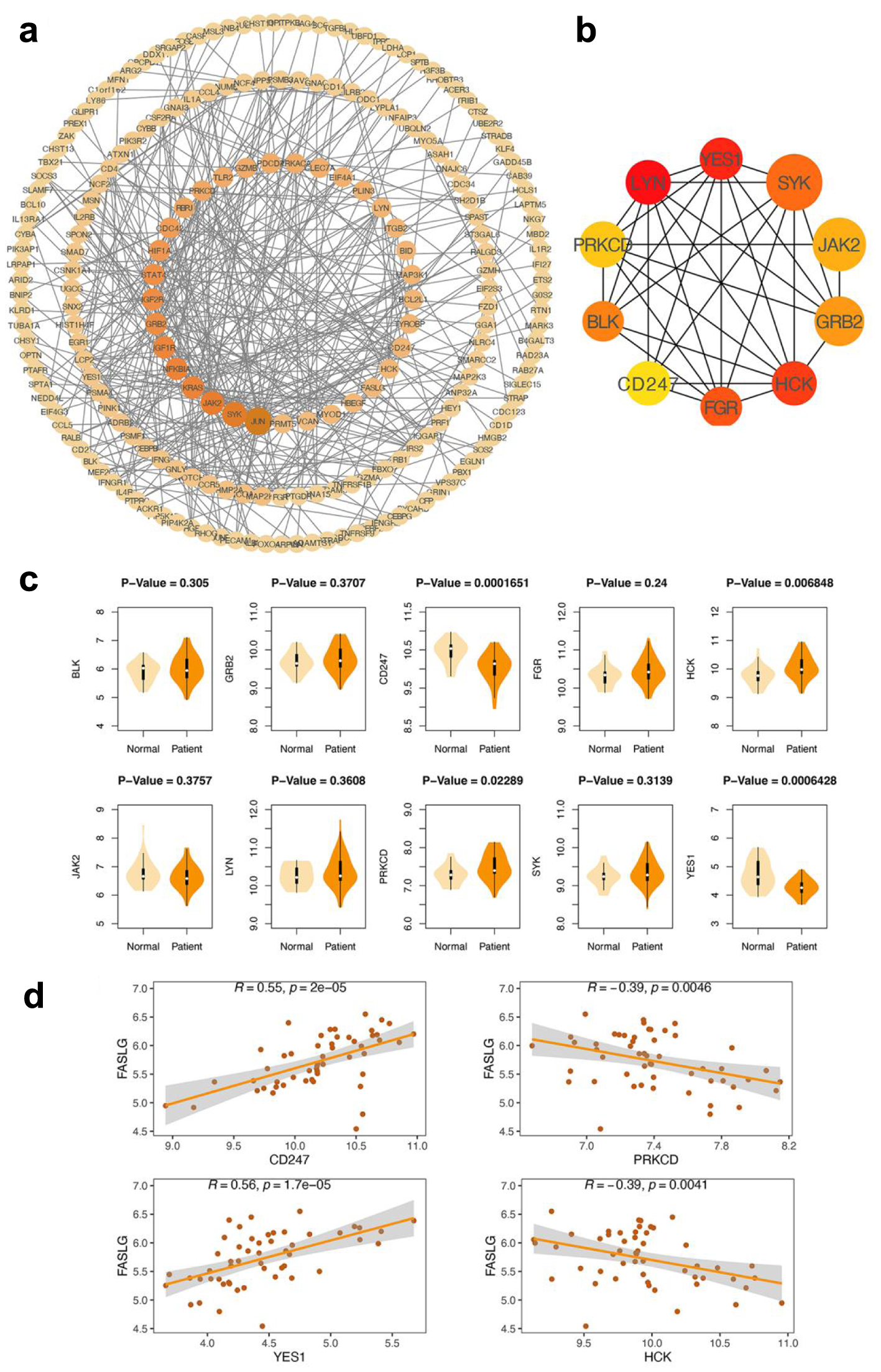 Click for large image | Figure 5. Construction of PPI network and identification of hub genes. (a) PPI network. The larger the diameter of the circle and deeper the color, the greater its mesoscopic centrality indicated, which has a central position in the network. (b) CytoHubba screens for 10 central genes. A deeper color represents a higher MCC score. Genes that tend to obtain high MCC scores are key genes associated with FASLG. (c) Examination of the differences in expression of 10 central genes in the AMI and normal groups. YES1, HCK, CD247 and PRKCD showed significant differences between the AMI and normal groups (their P values were less than 0.05) and were considered to be involved in the myocardial infarction process. (d) Correlation analysis of YES1, HCK, CD247, PRKCD, and FASLG. R-values closer to 1 indicate a stronger positive correlation and r-values closer to -1 indicate a stronger negative correlation. AMI: acute myocardial infarction; PPI: protein-protein interaction. |
Correlation analysis of FASLG and hub genes with immune cell infiltration in patients with AMI
Significant differences were detected in the proportion of immune cells between individual samples in the AMI group using the CIBERSORT algorithm analysis (Fig. 6a). Two immune cell subtypes with characteristic genes unexpressed in the samples were removed. Further study revealed that eosinophils, natural killer (NK) cells resting, T cells CD4 memory activated, and T cells gamma delta presented differences in FASLG high expression group versus FASLG low expression group. Neutrophils and eosinophils showed differences in the FASLG high expression group versus the normal group. Monocytes, neutrophils, NK cells resting, T cells CD4 memory activated, and T cells gamma delta showed differences in the FASLG low expression group versus the normal group (Fig. 6b). Spearmen correlation analysis indicated that FASLG was positively correlated with NK cells resting, T cells CD4 memory activated, and T cells gamma delta, and negatively correlated with B cells naive and macrophages M0 (Fig. 6c). CD247 was positively correlated with T cells CD4 memory resting, T cells gamma delta, and negatively correlated with macrophages M0, neutrophils (Fig. 6d). YES1 was negatively correlated with T cells follicular helper only (Fig. 6e). The findings further indicated that the expression level of FASLG probably impacted the immune status of AMI patients.
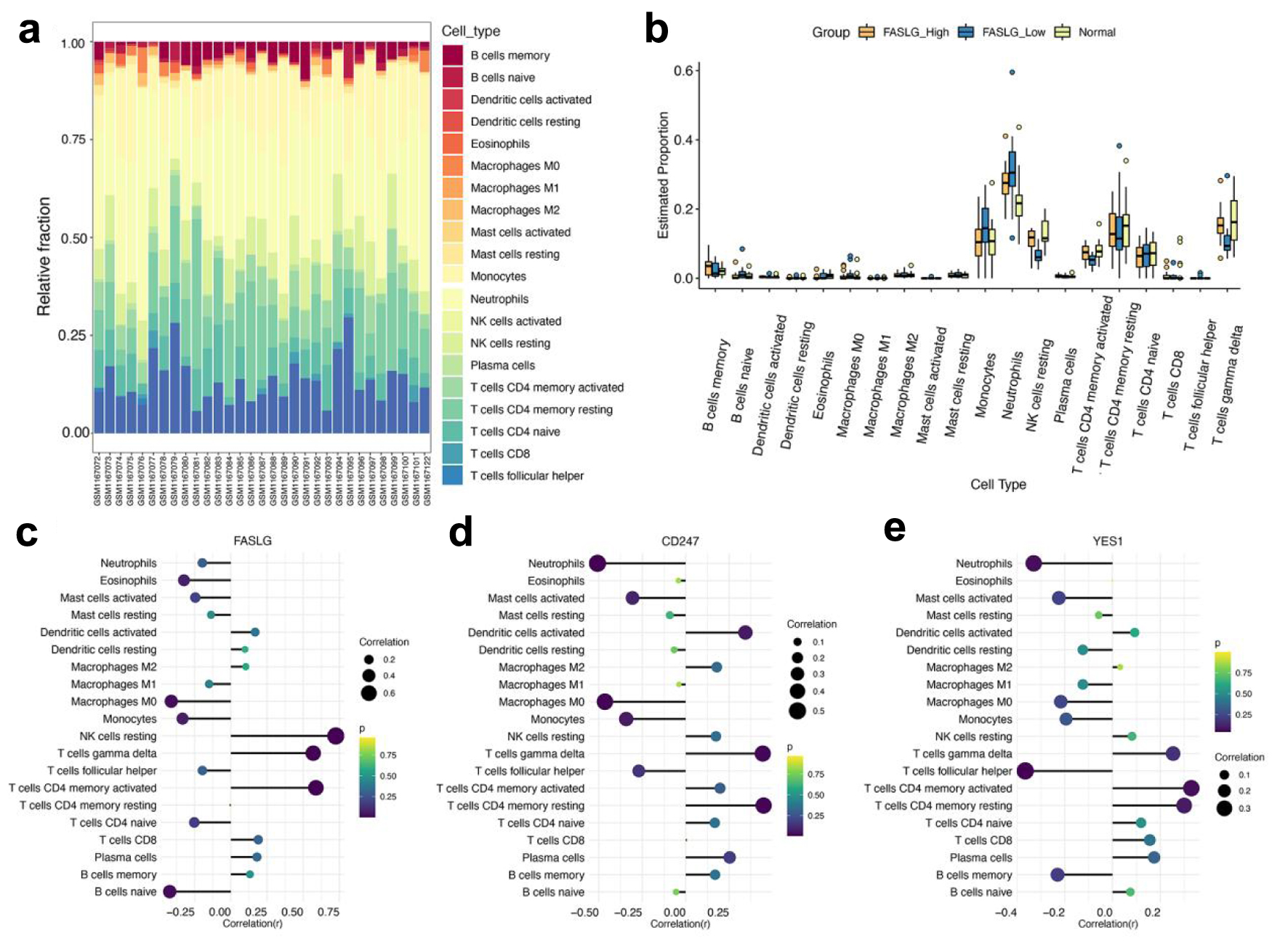 Click for large image | Figure 6. Analysis of immune cell infiltration assessment. (a) The box plot shows the relative percentages of different types of immune cells in all AMI samples. The horizontal axis shows the 31 AMI samples, and the length of each color block on the vertical axis indicates its percentage (0-1) of all immune cells. (b) Comparison of the proportion of various immune cell infiltrates in the FASLG high expression group, FASLG low expression group and normal group. (c-e) The correlations of FASLG, CD247, and YES1 with various immune cells were analyzed separately, and the larger circles and darker colors indicated their more obvious correlations. AMI: acute myocardial infarction. |
| Discussion | ▴Top |
The high mortality rate of AMI imposes a serious economic burden on society and families. Timely diagnosis and prediction of myocardial infarction contribute to improved prognosis of AMI. The currently used diagnostic indexes for AMI, such as cardiac troponin T (cTnT) and creatine kinase isoenzyme (CK-MB), still have limitations and might lead to underdiagnosis [25]. Recent studies have found that necrotizing apoptosis may participate in the regulation of cardiomyocyte death after the onset of AMI and affect ventricular remodeling after AMI. Upregulation of RIP1 and RIP3 after AMI activates the RIP1-RIP3-MLKL axis, which exacerbates cardiomyocyte death [26]. In addition, RIP3 can also cause necroptosis in cardiomyocytes through mobilization of Ca2+-calmodulin-dependent protein kinase (CaMKII) [27]. Therefore, the search for biomarkers associated with necroptosis to identify AMI patients is a feasible approach.
Blood samples from AMI patients and normal subjects were collected from the GEO database, and we found that FASLG and IFNG were genes associated with AMI, and necroptosis by WGCNA and differential analysis. FASLG (FAS ligand, also known as CD95L) is a member of the tumor necrosis factor (TNF) superfamily, and its binding to its receptor FAS could initiate apoptotic signaling causing apoptosis and immune response. It has been found that FASLG shows a down-regulation trend in AMI patients [28]. In addition, prostaglandin-E1 (PGE1) protects cardiomyocytes from I/R injury by regulating the miR-21-5p/FASLG axis [29]. IFNG encodes interferon-γ (IFN-γ), which activates macrophages and promotes Th1 responses, and plays a role in the inflammatory response after the onset of AMI [30]. Next, we applied statistical tests and found that the expression of FASLG was statistically different in the AMI group and the normal control group, but the expression of IFNG was not statistically different in the two groups. The ROC curve demonstrates the capability of FASLG to diagnose AMI effectively and was validated in the validation set GSE60993. IFNG has a role in AMI, and the non-differential status of IFNG presented by the above results might be due to an insufficient sample size. In the present study, we identified FASLG as a valid biomarker for AMI [31].
To further investigate the function and mechanism of action of FASLG, we applied to GO, KEGG and GSEA to do functional and pathway enrichment studies. GO enrichment analysis revealed that the genes were enriched in the regulation of T-cell activation, regulation of immune response signaling pathways, activation of cells involved in immune response, and regulation of lymphocyte proliferation; KEGG enrichment analysis showed that FASLG was involved in the regulation of NK cell-mediated cytotoxicity, B-cell receptor signaling pathways, Th1 and Th2 cell differentiation, Th17 cell differentiation and other pathways. The role of inflammatory response in AMI has been demonstrated. The early stages after the onset of AMI are characterized by a variety of immune cells (e.g., monocytes, macrophages, and lymphocytes) involved in the inflammatory response in the myocardium. After AMI, neutrophils are first recruited to the ischemic cardiac tissue, where they can contribute to the repair of the infarcted myocardium by spreading the inflammatory response to the surrounding myocardium through degradation and degranulation and promoting infiltration of other immune cells [32]. Monocytes, macrophages, lymphocytes, and NK cells were together involved in the transition from the pro-inflammatory response to the anti-inflammatory repair phase after AMI, promoting healing and scar repair of the infarcted myocardium and preventing cardiac rupture [33]. T lymphocytes could exacerbate cardiomyocyte death by releasing cytokines such as IFN-γ and IL-17 [34]. Mature B lymphocytes selectively produce the chemokine CCL7, leading to increased myocardial injury and deterioration of cardiac function [35]. NK cells interact with macrophages, thereby enhancing each other’s activity and increasing the inflammatory response at the infarct site [36]. These results suggest that FASLG may participate in the development of AMI by modulating the immune response.
Next, we constructed a PPI network to obtain two central genes: CD247 and YES1 as potential downstream regulators of FASLG. CD247 (also known as CD3 zeta chain), an important component of the T-cell receptor (TCR)-CD3, is involved in the regulation of the immune response and functions in a variety of diseases with an inflammatory response involved (e.g., systemic sclerosis, type 2 diabetes) [37-39]. A previous study showed that FASLG, together with CD247, was identified as a potential biological target for MI immunotherapy [40]. It also has been noted that apoptosis mediated by FASL-expressing tumor cells in ovarian cancer can induce loss of CD3-zeta and CD3-ε chain expression in T lymphocytes [41]. YES1 is a tyrosine kinase [42], and it has been suggested that overexpression of miR-140 could inhibit cardiomyocyte apoptosis by regulating YES1, thereby reducing myocardial I/R injury [43]. In hepatocytes, NADPH oxidase activation induced by CD95L could lead to rapid activation of YES, which induced apoptosis [44]. These studies suggest that CD247 and YES1 might be involved in the development of AMI, and the exact mechanisms remain to be elucidated in future studies.
Finally, we investigated the infiltration of immune cells in AMI and their correlation with key genes using CIBERSORT. We found that neutrophils, NK cells resting, T-cell CD4 memory activation, and T-cell γδ infiltration in the AMI group showed differences from the control group, which could be related to the occurrence of inflammation after AMI. In addition, we found that both FASLG and CD247 correlate with T-cell γδ and macrophage M0; therefore, FASLG might affect T-cell γδ and macrophage M0 through regulation of CD247 to participate in the immune response. T-cell follicular helper cells play a key role in protective immunity by helping B cells produce antibodies against foreign pathogens [45]. In the KEGG enrichment analysis, we found that FASLG is involved in the B-cell receptor signaling pathway; therefore, FASLG might affect T-cell follicular helper cells by regulating YES1, and thus participate in the regulation of the B-cell receptor signaling pathway. The exact evidence requires further research to obtain. It has long been established that the complex inflammatory response after AMI is mediated by a combination of immune cells. Notably, the relationship between necrotizing apoptosis and the immune response is still being explored. It has been suggested that necrotizing apoptosis leads to the release of large amounts of intracellular substances after the disruption of cell membranes leading to the occurrence of inflammatory responses. Necrotizing apoptosis is an important modality leading to myocardial cell death after myocardial infarction, which in turn is closely associated with multiple immune responses and immune cells. It is reasonable to believe that necrotizing apoptosis may be involved in the development of AMI by regulating immune infiltration.
The main biomarker currently used for AMI detection is troponin, and despite its high specificity and sensitivity, there are certain limitations, such as troponin levels rise significantly only a few hours after the onset of AMI, which may lead to a delay in early diagnosis; certain non-cardiac diseases (e.g., chronic kidney disease, sepsis) may also lead to an increase in troponin levels, which affects the accuracy of the diagnosis [46]. Our study showed that FASLG showed a tendency to increase in the early stages after the onset of AMI, which is important for the early diagnosis and timely treatment of AMI. FASLG, as a complementary marker, in combination with markers such as troponin, can improve the accuracy and timeliness of the diagnosis. In conclusion, we identified FASLG as a possible biomarker for AMI based on the bioconductivity analysis method according to the flow chart in Figure 7a and explored the possible mechanisms of its involvement in the development of AMI (Fig. 7b), providing a new perspective for the early diagnosis of AMI.
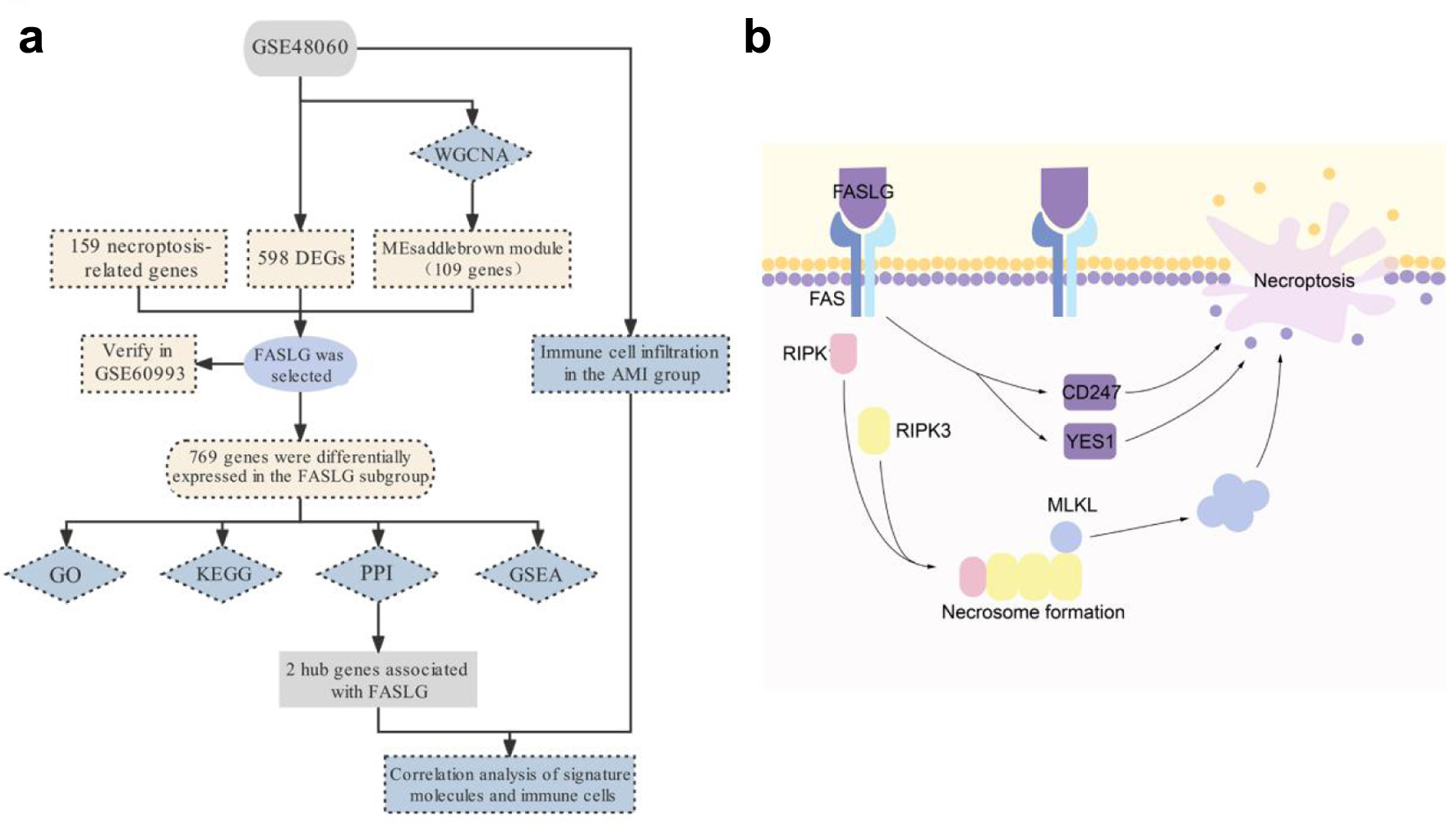 Click for large image | Figure 7. Schematic diagram of the workflow (a) and mechanism (b) of this study. |
Acknowledgments
We thank Professor Jianni Qi for her guidance on the design and writing of this study.
Financial Disclosure
This work was supported in part by grants from the Natural Science Foundation of Shandong Province (ZR2022MH146) and Shandong First Medical University Youth Science Foundation (202201-073).
Conflict of Interest
The authors declare that they have no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
HJB designed this article. HMJ and FXA collected and assembled the data. HMJ, FXA, MZY, YZ and HJB analyzed and interpreted. HMJ and FXA wrote the main manuscript text. YZ and HJB revised the manuscript. All authors reviewed and approved the manuscript.
Data Availability
The datasets used and/or analyzed during the current study are available from the first and corresponding author on reasonable request.
| References | ▴Top |
- Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet. 2017;389(10065):197-210.
doi pubmed - Gulati R, Behfar A, Narula J, Kanwar A, Lerman A, Cooper L, Singh M. Acute myocardial infarction in young individuals. Mayo Clin Proc. 2020;95(1):136-156.
doi pubmed - Bhatt DL, Lopes RD, Harrington RA. Diagnosis and treatment of acute coronary syndromes: a review. JAMA. 2022;327(7):662-675.
doi pubmed - Giannitsis E, Katus HA. Cardiac troponin level elevations not related to acute coronary syndromes. Nat Rev Cardiol. 2013;10(11):623-634.
doi pubmed - Li S, Wang A, Zhang Y, Tian X, Meng X, Wang Y, Li H, et al. Creatine kinase is associated with recurrent stroke and functional outcomes of ischemic stroke or transient ischemic attack. J Am Heart Assoc. 2022;11(6):e022279.
doi pubmed pmc - Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147(3):126-132.
doi pubmed - Aziz H, Zaas A, Ginsburg GS. Peripheral blood gene expression profiling for cardiovascular disease assessment. Genomic Med. 2007;1(3-4):105-112.
doi pubmed pmc - Wu Y, Jiang T, Hua J, Xiong Z, Chen H, Li L, Peng J, et al. Integrated bioinformatics-based analysis of hub genes and the mechanism of immune infiltration associated with acute myocardial infarction. Front Cardiovasc Med. 2022;9:831605.
doi pubmed pmc - Wang Y, Zhang X, Duan M, Zhang C, Wang K, Feng L, Song L, et al. Identification of potential biomarkers associated with acute myocardial infarction by weighted gene coexpression network analysis. Oxid Med Cell Longev. 2021;2021:5553811.
doi pubmed pmc - Zhao E, Xie H, Zhang Y. Predicting diagnostic gene biomarkers associated with immune infiltration in patients with acute myocardial infarction. Front Cardiovasc Med. 2020;7:586871.
doi pubmed pmc - Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11(10):700-714.
doi pubmed - Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2010;22(2):263-268.
doi pubmed pmc - Oerlemans MI, Liu J, Arslan F, den Ouden K, van Middelaar BJ, Doevendans PA, Sluijter JP. Inhibition of RIP1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Res Cardiol. 2012;107(4):270.
doi pubmed - Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, Kirsch P, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477(7364):330-334.
doi pubmed - Lin J, Li H, Yang M, Ren J, Huang Z, Han F, Huang J, et al. A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Rep. 2013;3(1):200-210.
doi pubmed - Park HH, Kim HR, Park SY, Hwang SM, Hong SM, Park S, Kang HC, et al. RIPK3 activation induces TRIM28 derepression in cancer cells and enhances the anti-tumor microenvironment. Mol Cancer. 2021;20(1):107.
doi pubmed pmc - Chang L, Wang Z, Ma F, Tran B, Zhong R, Xiong Y, Dai T, et al. ZYZ-803 Mitigates Endoplasmic Reticulum Stress-Related Necroptosis after Acute Myocardial Infarction through Downregulating the RIP3-CaMKII Signaling Pathway. Oxid Med Cell Longev. 2019;2019:6173685.
doi pubmed pmc - Gao X, Zhang H, Zhuang W, Yuan G, Sun T, Jiang X, Zhou Z, et al. PEDF and PEDF-derived peptide 44mer protect cardiomyocytes against hypoxia-induced apoptosis and necroptosis via anti-oxidative effect. Sci Rep. 2014;4:5637.
doi pubmed pmc - Qin D, Wang X, Li Y, Yang L, Wang R, Peng J, Essandoh K, et al. MicroRNA-223-5p and -3p cooperatively suppress necroptosis in ischemic/reperfused hearts. J Biol Chem. 2016;291(38):20247-20259.
doi pubmed pmc - https://www.ncbi.nlm.nih.gov/geo/.
- Suresh R, Li X, Chiriac A, Goel K, Terzic A, Perez-Terzic C, Nelson TJ. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J Mol Cell Cardiol. 2014;74:13-21.
doi pubmed pmc - Park HJ, Noh JH, Eun JW, Koh YS, Seo SM, Park WS, Lee JY, et al. Assessment and diagnostic relevance of novel serum biomarkers for early decision of ST-elevation myocardial infarction. Oncotarget. 2015;6(15):12970-12983.
doi pubmed pmc - https://www.kegg.jp/.
- https://string-db.org/.
- Rittoo D, Jones A, Lecky B, Neithercut D. Elevation of cardiac troponin T, but not cardiac troponin I, in patients with neuromuscular diseases: implications for the diagnosis of myocardial infarction. J Am Coll Cardiol. 2014;63(22):2411-2420.
doi pubmed - Zhang H, Yin Y, Liu Y, Zou G, Huang H, Qian P, Zhang G, et al. Necroptosis mediated by impaired autophagy flux contributes to adverse ventricular remodeling after myocardial infarction. Biochem Pharmacol. 2020;175:113915.
doi pubmed - Zhang T, Zhang Y, Cui M, Jin L, Wang Y, Lv F, Liu Y, et al. CaMKII is a RIP3 substrate mediating ischemia- and oxidative stress-induced myocardial necroptosis. Nat Med. 2016;22(2):175-182.
doi pubmed - Wu K, Zhao Q, Li Z, Li N, Xiao Q, Li X, Zhao Q. Bioinformatic screening for key miRNAs and genes associated with myocardial infarction. FEBS Open Bio. 2018;8(6):897-913.
doi pubmed pmc - Tang M, Pan H, Zheng Z, Guo Y, Peng J, Yang J, Luo Y, et al. Prostaglandin E1 protects cardiomyocytes against hypoxia-reperfusion induced injury via the miR-21-5p/FASLG axis. Biosci Rep. 2019;39(12):BSR20190597.
doi pubmed pmc - Steppich BA, Moog P, Matissek C, Wisniowski N, Kuhle J, Joghetaei N, Neumann FJ, et al. Cytokine profiles and T cell function in acute coronary syndromes. Atherosclerosis. 2007;190(2):443-451.
doi pubmed - Ong SB, Hernandez-Resendiz S, Crespo-Avilan GE, Mukhametshina RT, Kwek XY, Cabrera-Fuentes HA, Hausenloy DJ. Inflammation following acute myocardial infarction: Multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol Ther. 2018;186:73-87.
doi pubmed pmc - Frangogiannis NG. Interleukin-1 in cardiac injury, repair, and remodeling: pathophysiologic and translational concepts. Discoveries (Craiova). 2015;3(1):e41.
doi pubmed pmc - Horckmans M, Ring L, Duchene J, Santovito D, Schloss MJ, Drechsler M, Weber C, et al. Neutrophils orchestrate post-myocardial infarction healing by polarizing macrophages towards a reparative phenotype. Eur Heart J. 2017;38(3):187-197.
doi pubmed - Yan X, Shichita T, Katsumata Y, Matsuhashi T, Ito H, Ito K, Anzai A, et al. Deleterious effect of the IL-23/IL-17A axis and gammadeltaT cells on left ventricular remodeling after myocardial infarction. J Am Heart Assoc. 2012;1(5):e004408.
doi pubmed pmc - Zouggari Y, Ait-Oufella H, Bonnin P, Simon T, Sage AP, Guerin C, Vilar J, et al. B lymphocytes trigger monocyte mobilization and impair heart function after acute myocardial infarction. Nat Med. 2013;19(10):1273-1280.
doi pubmed pmc - Knorr M, Munzel T, Wenzel P. Interplay of NK cells and monocytes in vascular inflammation and myocardial infarction. Front Physiol. 2014;5:295.
doi pubmed pmc - Call ME, Wucherpfennig KW. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol Immunol. 2004;40(18):1295-1305.
doi pubmed pmc - Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, Coenen MJ, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42(5):426-429.
doi pubmed pmc - Eldor R, Klieger Y, Sade-Feldman M, Vaknin I, Varfolomeev I, Fuchs C, Baniyash M. CD247, a novel T cell-derived diagnostic and prognostic biomarker for detecting disease progression and severity in patients with type 2 diabetes. Diabetes Care. 2015;38(1):113-118.
doi pubmed - Zhang L, Wang Q, Xie X. Identification of biomarkers related to immune cell infiltration with gene coexpression network in myocardial infarction. Dis Markers. 2021;2021:2227067.
doi pubmed pmc - Rabinowich H, Reichert TE, Kashii Y, Gastman BR, Bell MC, Whiteside TL. Lymphocyte apoptosis induced by Fas ligand- expressing ovarian carcinoma cells. Implications for altered expression of T cell receptor in tumor-associated lymphocytes. J Clin Invest. 1998;101(11):2579-2588.
doi pubmed pmc - Lun XK, Szklarczyk D, Gabor A, Dobberstein N, Zanotelli VRT, Saez-Rodriguez J, von Mering C, et al. Analysis of the human kinome and phosphatome by mass cytometry reveals overexpression-induced effects on cancer-related signaling. Mol Cell. 2019;74(5):1086-1102.e1085.
doi pubmed pmc - Yang S, Li H, Chen L. MicroRNA-140 attenuates myocardial ischemia-reperfusion injury through suppressing mitochondria-mediated apoptosis by targeting YES1. J Cell Biochem. 2019;120(3):3813-3821.
doi pubmed - Reinehr R, Becker S, Eberle A, Grether-Beck S, Haussinger D. Involvement of NADPH oxidase isoforms and Src family kinases in CD95-dependent hepatocyte apoptosis. J Biol Chem. 2005;280(29):27179-27194.
doi pubmed - Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185-189.
doi pubmed pmc - Park KC, Gaze DC, Collinson PO, Marber MS. Cardiac troponins: from myocardial infarction to chronic disease. Cardiovasc Res. 2017;113(14):1708-1718.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cardiology Research is published by Elmer Press Inc.


